Announcing our first application: Therapeutics manufacturing.
What if we could manufacture advanced therapies better, faster, and cheaper by turning it into a quantitative engineering problem? We are integrating synthetic biology and computer-aided design to engineer more productive molecular factories for biologics, viral vectors, and RNA. Our approach couples standardized host cell lines, libraries of characterized genetic parts, and computational models to control expression, secretion, stability, post-translational modifications, and beyond. Contact us to learn more.
Biotech needs better design tools
Imagine an alternate universe where rockets were designed not through careful engineering, but instead via trial-and-error: by using random combinations of components, no theoretical understanding of propulsion, and a design strategy that boils down to crossing your fingers. It would be disastrous! Almost every launch would end in a fireball, and on rare occasions one would achieve lift-off. Believe it or not, this is similar to how most genetic engineering is done today.
Don’t get me wrong – over the past half-century, genetic engineering has unquestionably transformed the world for the better. It has given us more efficacious therapeutics, more productive crops, and more sustainable chemicals. But despite the name, “genetic engineering” is not yet an engineering discipline. Even after decades of research, the process of genetically modifying a cell is driven by trial-and-error. For any particular application, anywhere from dozens to thousands of different genetic designs are tested because it’s hard to know in advance which, if any, will work. The robust design and modeling tools that exist for other industries do not exist for biology.
To be fair, this process actually works remarkably well in certain cases. In fact, evolution does a version of this in nature. By taking the reins on this process in a controlled laboratory setting (i.e., directed evolution), a powerful method for developing enzymes (and more) emerges. But you need some starting activity to optimize from. If your target is a complex, multi-part system that introduces entirely new functions to a cell, the starting point becomes even more essential. Evolution can optimize a system efficiently, but good initial design is a prerequisite to the process.
Currently, the design of complex multi-part genetic systems is often a shot in the dark. It’s revealing that most biotech organizations focus heavily on high-throughput screening – it’s an attempt to brute force the problem; to side-step the tooling gap. This is like hoping to stumble upon the solution, and it becomes less tractable as the complexity of a genetic system increases. The result is the current state of biotechnology: protracted product development timelines, sky-high R&D costs, and an inability to realize more ambitious ideas.
Brute forcing genetic design will not scale as the field advances. We need better tools that can help us take more intelligent shots on goal when designing a living system. The development process needs to be more reliable, more like programming a cell, than seeing what sticks through trial-and-error.
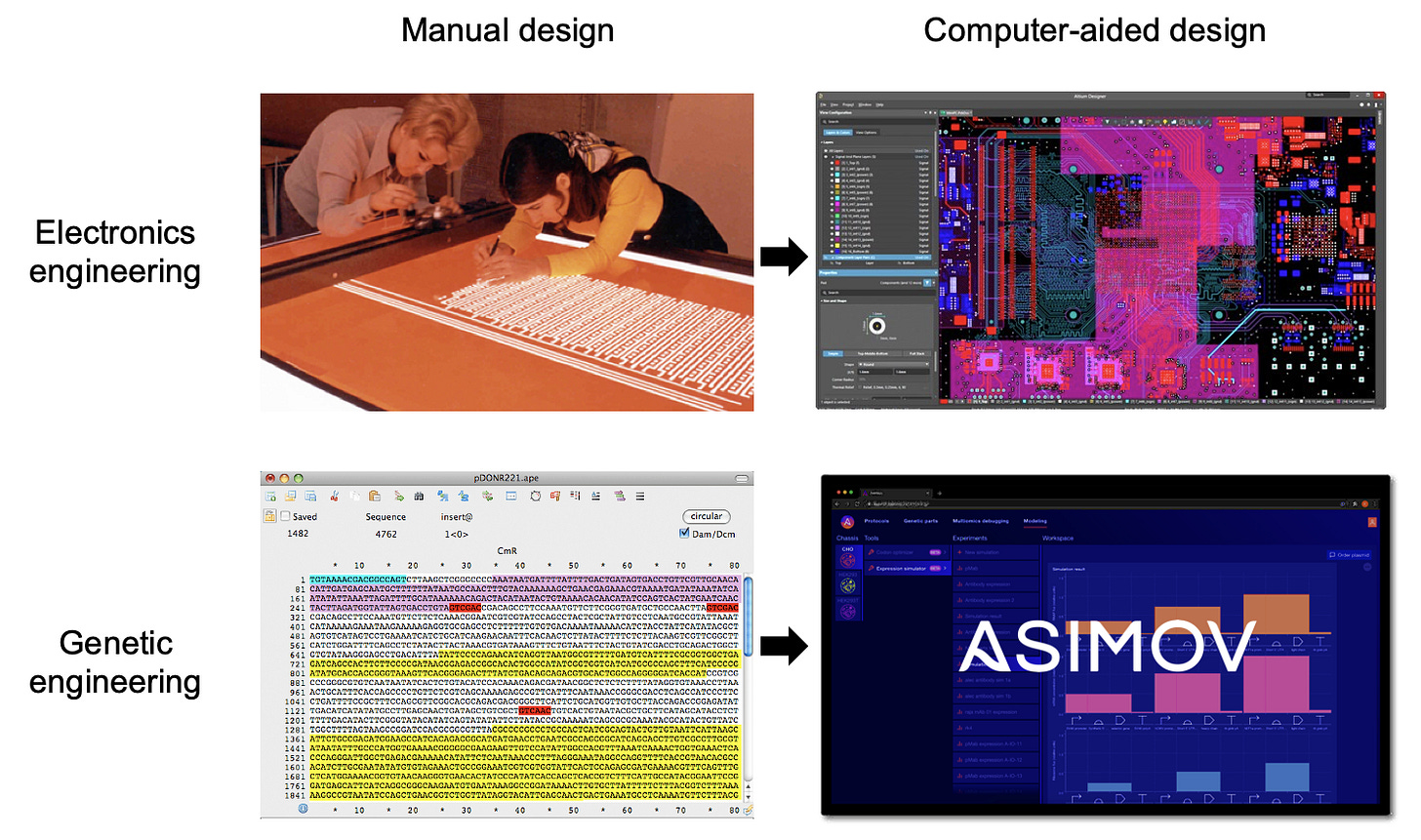
This is a core goal of synthetic biology, but the field's tools are still too primitive. Strikingly, the most widely used software in the field is a DNA sequence editor - a simple app to visualize and annotate strings of A’s, T’s, G’s, and C’s. A DNA sequence editor isn’t design software; it’s a word processor with most of the alphabet missing!
Compare that to the sophisticated software used in other engineering disciplines, many of which have matured over the same timeframe as genetic engineering. Look at the tools used by engineers to design skyscrapers, integrated circuits, and indeed rockets. We do not stumble upon these via trial-and-error; we forward engineer them with powerful CAD tools.
Biology isn’t rocket science – it’s harder. Biology is bewilderingly complex at every level, and we need better tools to design it. At Asimov, we are tackling this problem head on by coupling computer-aided design, data-driven models, and experimental synthetic biology.
Introducing Kernel: A CAD platform for biology
To achieve this vision, we’re building Kernel, a cloud-based software suite for genetic designers that is coupled with real biological components and data-driven models. It’s the world’s first fully-integrated, commercial CAD platform for biology. Our goal is to enable quantitative design, simulation, and debugging of genetic systems through a user-friendly interface. It's an attempt to move far beyond the DNA sequence editor. An early version of our platform is in the hands of customers today, and we will be launching the next major version later this year.
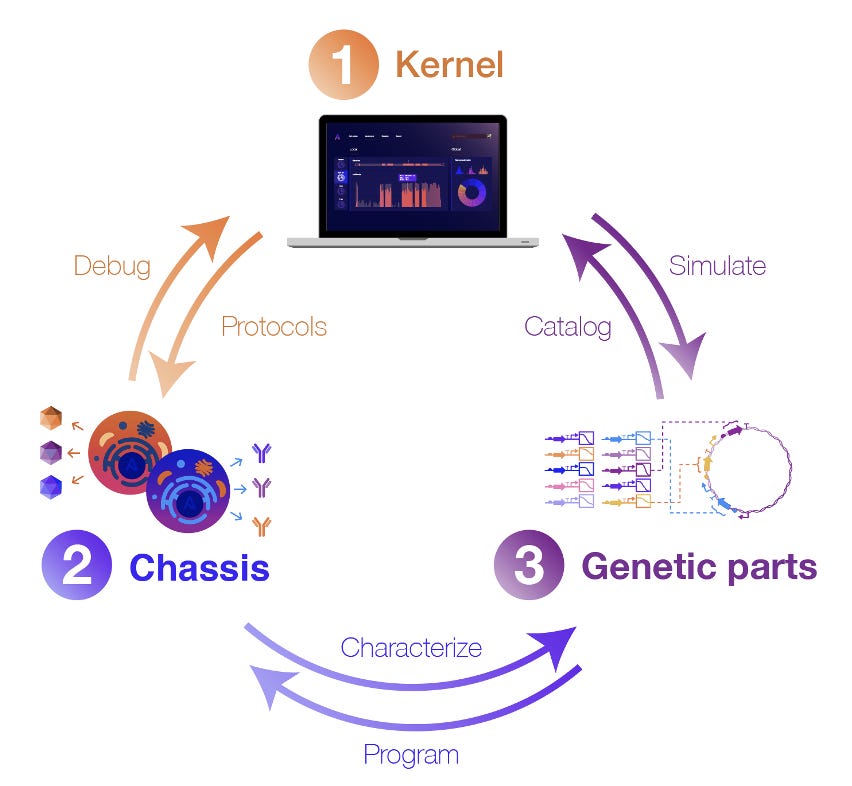
But software alone isn’t enough. To reliably predict cellular behavior, software needs to be deeply connected to experimental data and validated biology. As such, Kernel development is integrated with optimized host cell lines and genetic parts that encode an array of cellular functions. We believe that tight integration of digital and biological components has the potential to transform how genetic design is done.
Genetic design using professional parts
Genetic systems are designed using DNA sequences called genetic parts that encode cellular functions. Parts are composed together to create more complex genetic designs.
A surprisingly small roster of parts has formed the backbone of biotechnology since the field’s beginnings – parts like GFP (green fluorescent protein), CMV promoter, ColE1 origin of replication, and more recently CRISPR-Cas9. Although the concept of characterized parts has been around for decades, what currently exists barely scratches the surface of all possible useful parts. There’s an unmet need for high-quality parts to control diverse aspects of cell biology, with quantitative data to guide their use.
Today, most genetic constructs are designed in a relatively ad hoc fashion: a minimal set of legacy parts (often with performance issues) is frequently used for all genetic engineering projects within an organization. In cases where those legacy parts don’t suffice, parts need to be onboarded by scraping research papers and copying DNA sequences from the supplemental PDF (hopefully the formatting doesn't get messed up! I've been burned by that more than once). Often, these parts do not have the necessary quality of data/metadata to get things working in a new setting. This is an inefficient engineering process, and it contributes to the punishing timelines and R&D costs of biotech products.
To address these limitations, our team designs and curates thousands of useful genetic parts, devices, and integrated systems. These parts are developed and extensively characterized in our lab, and then the corresponding performance data is codified into Kernel, where it can be accessed by users. Crucially, we characterize these genetic parts in a specific set of optimized host cell lines (called a “chassis” in synthetic biology parlance). By studying how each part behaves in a specific chassis, the cell line becomes a more controlled environment, meaning new genetic systems are less likely to fail than if they were installed in an entirely different host cell.
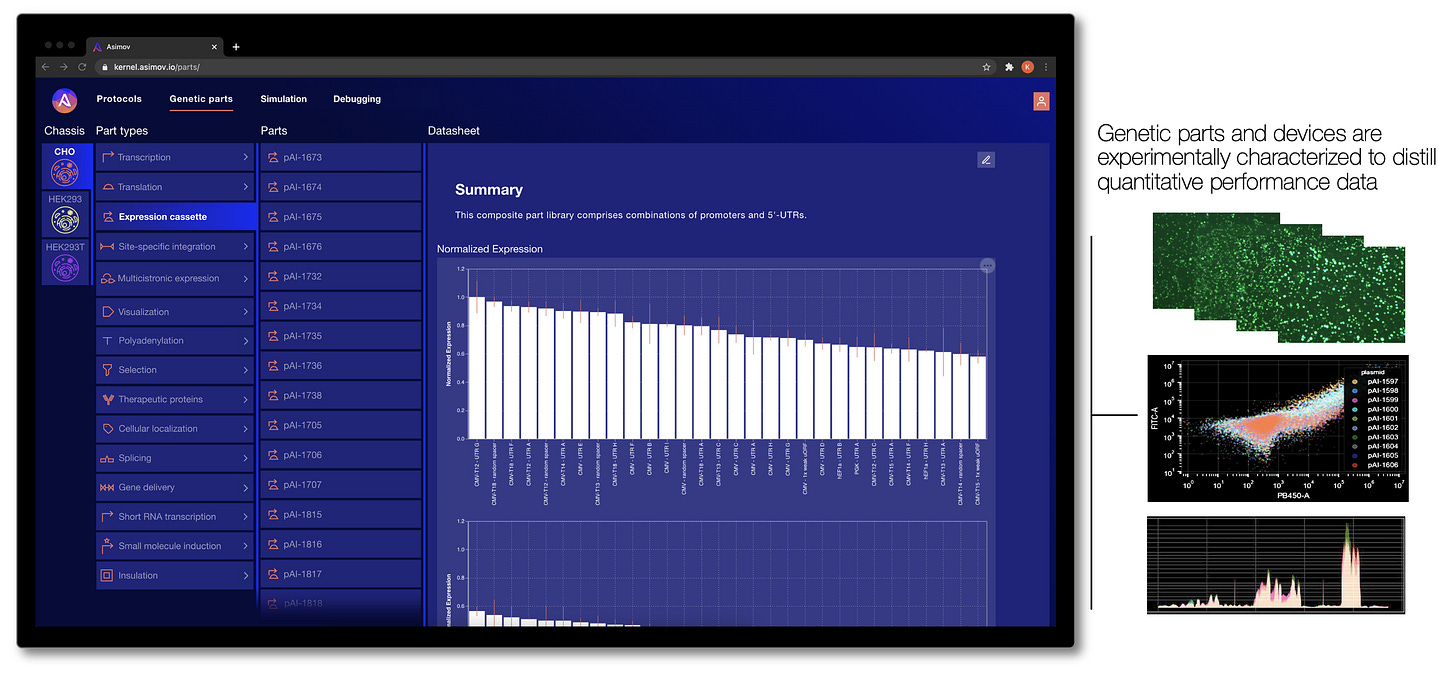
How do we decide what parts to develop for Kernel? It's simple - we work backward from the big challenges in the industry by talking to customers. It’s product management at the genetic level. We’re continuously growing our library of genetic parts, with a current focus on functions related to therapeutic design and production (e.g., tissue specific expression, epigenetic insulation, transposase integration, small-molecule induction, protein secretion, sub-cellular localization, and more).
One day, we strive to have cataloged millions of characterized parts for all types of biotech application. Our goal is to provide a comprehensive set of engineering-grade parts for the entire biotech community. But we don't want to stop at a parts catalog - we aim for this high-quality characterization data to enable something previously impossible: predictive simulation.
Data-driven simulations of biology
One of the frustrations of designing a genetic system today is that it typically relies on intuition: the engineer forms a mental model of how the system functions. This isn’t quantitative, and it fails completely for systems of even moderate complexity. To address this gap, we have taken inspiration from other engineering disciplines, such as computational fluid dynamics, finite element analysis, and electrical circuit models to build a modeling stack for biology. We develop many different types of models at Asimov, from those used to predict splicing, signal peptide cleavage, codon optimization, cellular metabolism, and bioreactor processes. Below, we highlight one such model called the Genetic Simulator.
Given a specific cell chassis and a genetic system designed in Kernel, the Genetic Simulator makes predictions about the biophysical behavior of the system. Through Kernel's interface, a genetic design can be refined until the predictions look promising, and then the DNA sequence for that genetic program can be requested directly. (We fulfill custom plasmid requests by coordinating external DNA synthesis and internal assembly, and then we ship the prepped plasmid to the user.)
The Genetic Simulator was initially developed as part of the DARPA ASKE program. It is a hybrid machine learning and physics-based model that accepts custom genetic designs as input. Multiple levels of cellular phenomena are simulated through time for each component part, including ligand sensing, regulation, transcription, translation, and more.
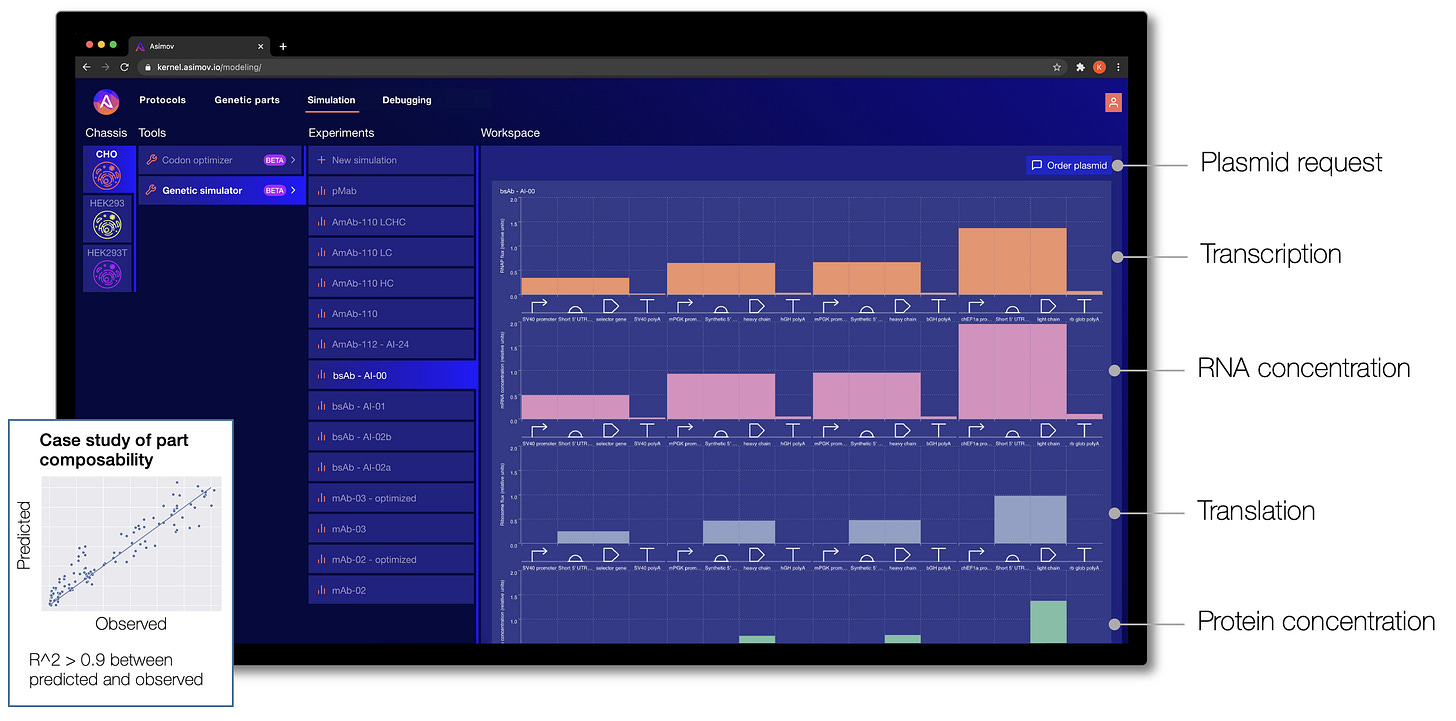
This is just the beginning. We plan to incorporate other phenomena critical for cell design, including post-translational modifications, organelle trafficking, and bioreactor process models.
Simulation of biological systems is arguably the hardest problem in the world today. Importantly, simulation doesn't need to be perfect; it just needs to be good enough to help triage an unimaginably large design space. There are many places where our models currently falter: contextual effects, parts interference, metabolic burden, resource competition, and so on. But we believe these problems are solvable by approaching them from both ends: by intentionally developing more modular/insulated systems and figuring out the design rules (in other words, 'engineering a biology that is more engineerable') and by continuously improving the models with new data.
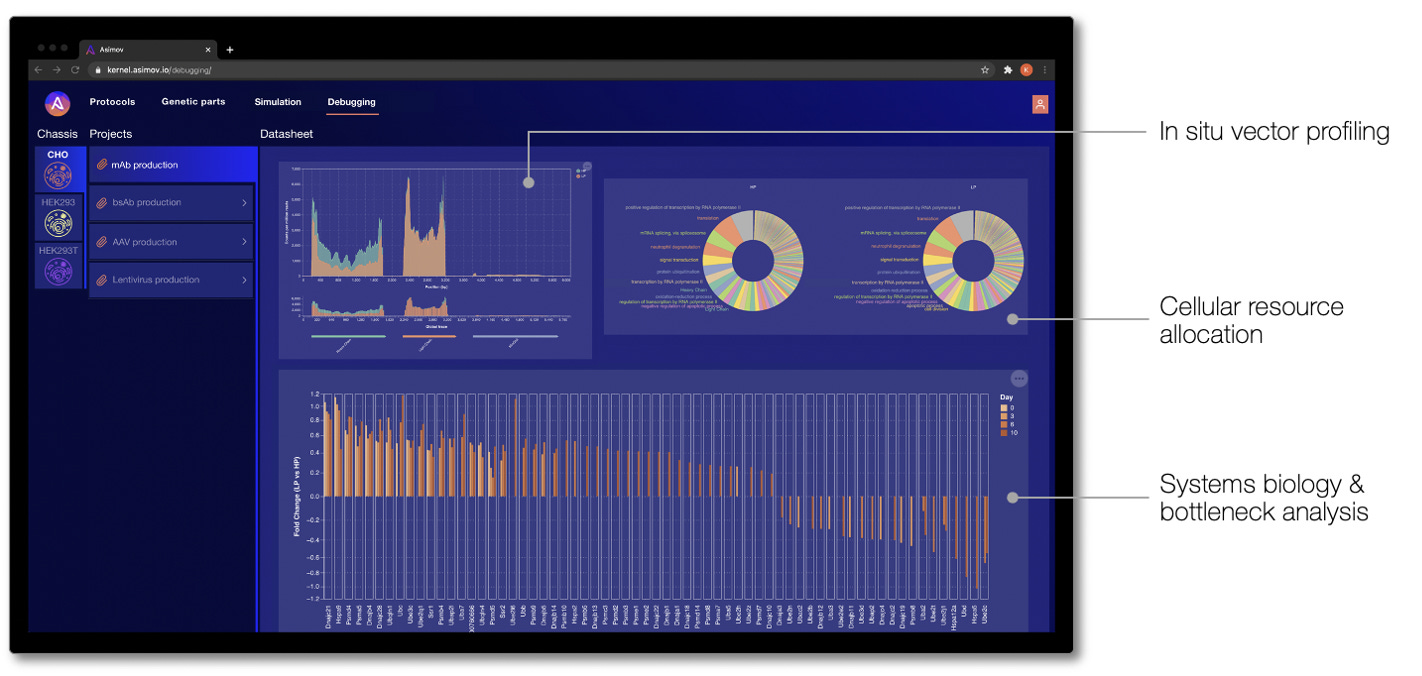
Debugging cell behavior to learn from and improve genetic designs
In other CAD domains, when a system behaves unexpectedly, engineers have powerful “debuggers” to diagnose what went wrong. Unfortunately, when an engineered cell behaves unexpectedly, it’s difficult to know what went wrong and how to fix it. To address this, we built a simple UI in Kernel to communicate -omics debugging results to users. While in nascent form, these analyses focus on a core set of standard bioinformatics measurements to understand a genetic system and its broader effects. Where the Genetic Simulator predicts how a genetic system behaves before it's physically built, debugging tells you what's actually happening 'under the hood' of an engineered cell.
Analyses that we have reported to customers via Kernel include in situ genetic part performance, cellular resource allocation, and dysregulation of endogenous networks. Ultimately, the goal of debugging is to find the root cause of the problem and guide the design of a better genetic program that achieves the desired behavior.
Focus area: improving therapeutics manufacturing
To showcase the potential of a hybrid synthetic biology and computer-aided design approach, we identified an application area with high societal benefit, clear technical challenges with unmet need, and broad utility for many different products. Specifically, for our first application we are laser focused on the design and manufacture of biologics, viral vectors, and RNA.
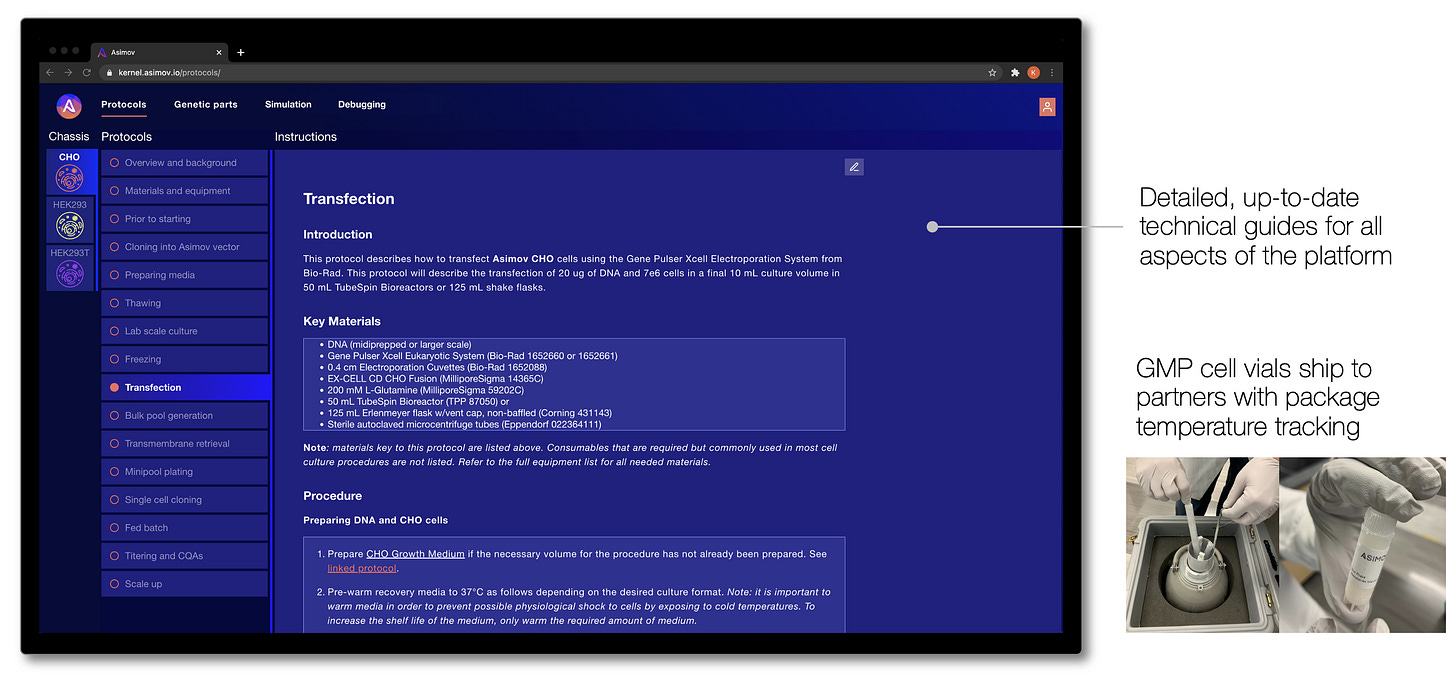
For these modalities, we have developed a set of optimized mammalian host cell chassis, genetic parts, and design tools for the clinical and commercial manufacture of these therapeutics. Host cell chassis are banked under CGMP (“current good manufacturing practice”, as required by the FDA) and provided to our partners with the associated regulatory documentation. Cells are shipped directly to our partners in liquid nitrogen and we grant them cloud-based access to Kernel and our genetic parts library and plasmid templates.
For commercial-grade biologics production, we have developed high performance glutamine synthetase-knockout Chinese hamster ovary (CHO) cells for clinical and commercial production of monoclonal antibodies, bispecific/multifunctional antibodies, and fusion proteins. We have also developed genome-edited variant CHO cells to control antibody glycosylation. These cell lines come with recommended genetic parts and plasmids to enable faster, transposase-mediated cell line development. For customer molecules in-house, we have seen substantial productivity and quality gains compared to their current platforms.
For cell and gene therapy, viral vector manufacturing is a huge bottleneck in the industry. We believe our platform has the potential to drive dramatic reductions in cost and improvements in quality. We provide our partners with GMP-banked HEK293 or HEK293T-derived cells for adeno-associated virus (AAV) and lentiviral vectors. We also provide access to genetic parts to engineer the payload, including in vivo-validated promoters for tissue specific expression.
All of these elements seamlessly integrate into standard industry cell line development and manufacturing workflows, and we provide detailed technical guides that walk users through each step of working with the cells and genetic systems. These technical guides are accessible via Kernel, which means our users always have the most up-to-date protocols to get the most out of the platform.
With over 25 early-access partners, including top-10 pharma companies, virtual biotechs, and leading CDMOs, we’re tackling multiple programs in AAV, lentivirus, and antibody production.
The future of genetic design
Building a CAD platform for biology has massive potential, but it is a monumental challenge. We are taking a careful, holistic approach that couples design software with characterized genetic parts and standardized host cell chassis.
In summary: Standardized chassis serve as a controlled environment for genetic part characterization. Genetic part characterization, in turn, feeds into models for complex system and bioprocess design. Multi-omics debugging enables a deeper understanding of the cell and drives actionable insight for the engineered system. Collectively, we believe these approaches have the potential to move biotech R&D past the rut of slow trial-and-error cycles. Each of these concepts has well-vetted roots in the development of Cello, an integrated software and synthetic biology platform for automating genetic logic circuit design.
While our current focus is therapeutics, we are building our approach in a general way that has the potential to be applied in other biotechnology domains, such as biomaterials, cellular agriculture, and specialty chemicals.
Our ultimate goal is to enable genetic designers to create biological systems that are as complex, subtle, and elegant as those created by nature. Long-term, we want to provide designers with powerful abstractions for specifying a cell’s behavior, whether it be programming an immune cell to sense and respond to cancer or dynamically regulating a cell’s metabolism in a bioreactor. In parallel we are working to automate the genetic implementation, thereby freeing the designer to focus on the high level functional, environmental, security, and safety requirements for their systems.