Central Dogma in 7 Experiments
A brief history of molecular biology’s seminal achievement.
In the days before DNA sequencing, high-powered microscopes, and molecular biology textbooks, decoding the finer workings of a living cell often required arduous experiments and clever speculation.
The history of molecular biology is rife with eccentric scientists who drummed up creative experiments to study unseen molecules, and then used deductive reasoning to piece a larger puzzle together. Mapping the Central Dogma is their crowning achievement.
The Central Dogma was first described by Francis Crick, the Cambridge scientist who solved DNA’s structure with James Watson, based on x-ray images obtained by Rosalind Franklin. In 1958, Crick wrote that once genetic information has passed into protein, “it cannot get out again.”
Although students typically learn the Central Dogma as something like DNA → RNA → protein, or “DNA is transcribed to RNA which is translated to protein,” this is not what Crick originally said. There are also exceptions to the oft-mentioned DNA→RNA→protein depiction; RNA is reverse transcribed into DNA, for example, and prions are protein aggregates that replicate themselves. Crick regretted naming his idea the ‘Central Dogma,’ he wrote in Nature, because the idea itself was speculative. Crick had misunderstood the definition of the word dogma.
Still, the ways in which cells read instructions encoded in DNA to create all the proteins necessary for life is the cornerstone of modern molecular biology. The scientists who cracked this code were often brilliant thinkers, and their experiments ought to be an inspiration for future genetic designers hoping to make discoveries in areas where we are currently most blind.
In this essay, we highlight 7 experiments that elucidated the Central Dogma and information processing in cells. These experiments include those that first isolated the intermediate molecule between DNA and proteins, called messenger RNA, cracked the genetic code, and solved the basic mechanism for DNA replication in living cells.
Experiments described in this essay are important, but not exhaustive. Biological knowledge is built up, slowly, by the collective efforts of hundreds of scientists. Only a book like The Eighth Day of Creation, by Horace Judson, could even begin to do justice to the rich and beautiful history of molecular biology. This essay focuses on a few important years, and is inspired by The Generalist’s article on the history of AI.
The Structure of DNA (1953)
Friedrich Miescher, a Swiss chemist, was the first person to isolate DNA. In 1869, he collected pus-covered bandages from patients at a university hospital and extracted a sticky substance from them. Miescher called this substance nuclein.
For decades after, most biologists believed that Miescher’s discovery was little more than a quaint curiosity. Early molecular biologists (the coin was termed in 1938) thought that proteins, rather than DNA, was the genetic material of living cells. Proteins are built from many different amino acids, and appear in all kinds of different shapes and sizes. This made them seem like the likelier option for genetic material.
By 1944, though, this view began to crumble when three scientists at the Rockefeller Institute in New York City, named Oswald Avery, Colin MacLeod, and Maclyn McCarty did an experiment to identify the molecule responsible for carrying genetic information. Their results pointed to DNA.
The trio isolated protein and DNA from different strains of Streptococcus pneumoniae, a virulent bacterium, and then used enzymes to break down the molecules. The digested proteins and DNA were inserted into a harmless strain of bacteria, and the scientists waited to see if either molecule would turn the harmless cells virulent.
When digested DNA was added to the cells, the harmless bacteria adopted the traits of the virulent strain, but this did not happen with digested proteins. These results suggested that DNA, and not proteins, was a carrier of hereditary information.
A few miles north, at Columbia University, a biochemist named Erwin Chargraff read the Avery-MacLeod-McCarty paper and was "deeply moved by the sudden appearance of a giant bridge between chemistry and genetics," as he later wrote. Chargaff had an academic background in molecular chemistry. He realized that, if DNA was indeed the genetic material, then perhaps a chemist could dissect how it differs across organisms and thus explain the rich diversity of the natural world.
Chargaff’s team spent several years chewing up DNA sequences, separating out the individual nucleotides on pieces of paper, and exposing the nucleotides to a UV spectrophotometer. They repeated this for DNA molecules harvested from yeast, bacteria, beef spleens, and calf thymus. By 1949, Chargaff had cracked a basic principle of the DNA code:
"The desoxypentose nucleic acids from animal and microbial cells contain varying proportions of the same four nitrogenous constituents, namely adenine, guanine, cytosine, thymine…Their composition appears to be characteristic of the species, but not of the tissue, from which they are derived."
In other words, Chargaff correctly determined that every organism on Earth uses DNA molecules that are made from the same four letters. Genetic material only differs, from one species to the next, by the order in which the four nucleotides appear. Chargaff also noted that "the molar ratios of total purines to total pyrimidines, and also of adenine to thymine and of guanine to cytosine, were not far from 1." Said another way, the amount of ‘A’ in DNA is always equal to the total amount of ‘T’. Ditto for ‘G’ and ‘C’ nucleotides.
Chargaff shared his results in a lecture at Cambridge University in 1952. Watson and Crick were in attendance. The following year, using x-ray diffraction images first obtained by Rosalind Franklin at King’s College London, and perhaps also Chargaff’s observations, Watson and Crick assembled a biophysically accurate model of DNA. Their model was made from crude, metal sheets, but clearly depicted a right-handed double-helix in which ‘A’ connects to ‘T’, and ‘G’ connects to ‘C’. The model was published in Nature on 25 April 1953.
Recent revelations have revised Rosalind Franklin’s role in solving DNA’s structure. In the classic telling of this tale, Franklin is “portrayed as a brilliant scientist, but one who was ultimately unable to decipher what her own data were telling her about DNA,” according to an article by Matthew Cobb & Nathaniel Comfort in Nature. “She supposedly sat on the image for months without realizing its significance, only for Watson to understand it at a glance.”
But this tale is not accurate. Newly unearthed documents, including a shelved article that Franklin wrote with Crick and Watson for Time magazine in 1953, now suggest that “Franklin did not fail to grasp the structure of DNA. She was an equal contributor to solving it.”
DNA Replication (1958)
Watson and Crick’s 1953 Nature paper concludes with one of the most famous passages in biology’s history:
“It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material.”
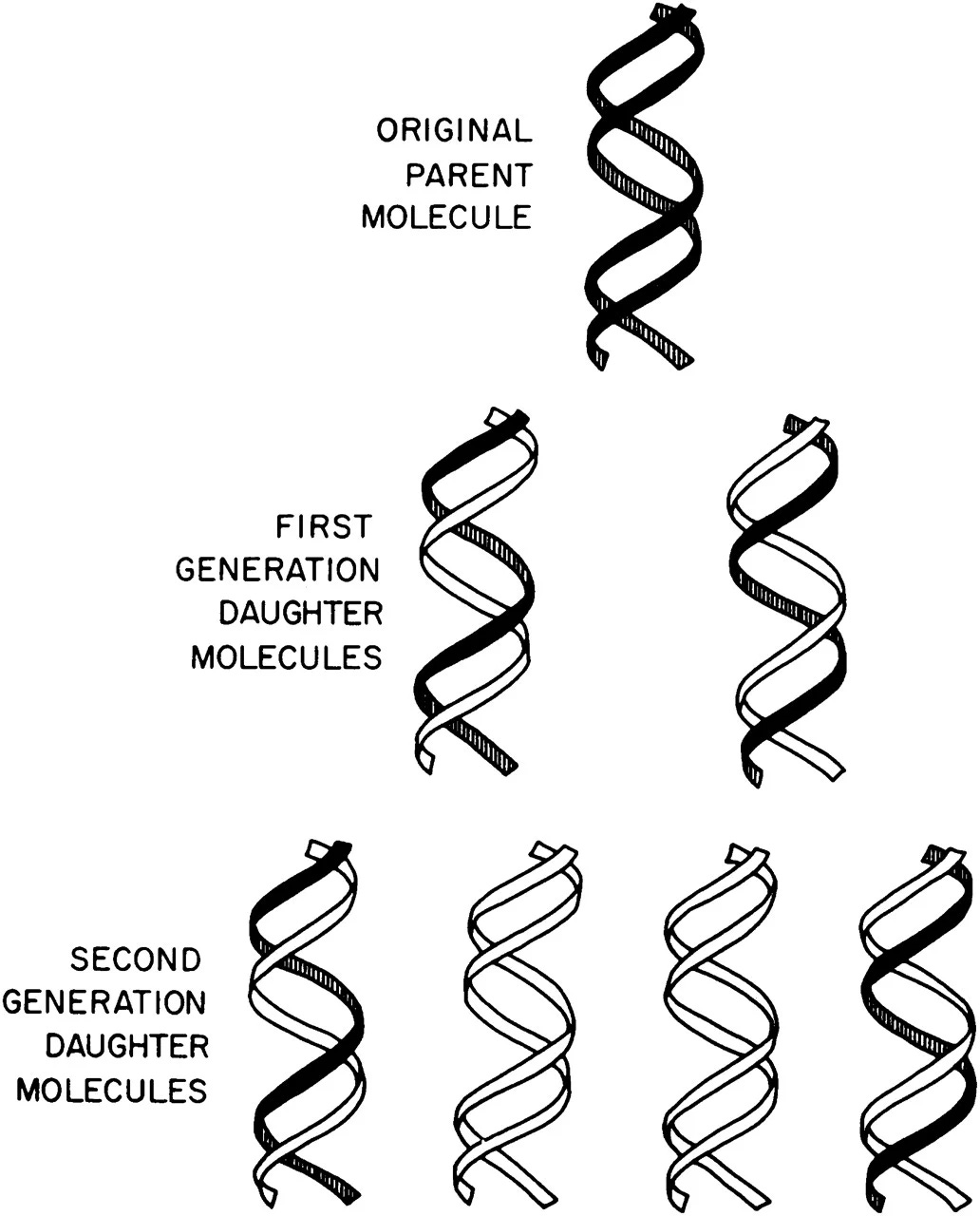
The Cambridge duo’s model correctly depicted DNA as a molecule composed from two interlocking strands, wherein ‘A’ always connects to ‘T’ and ‘G’ always connects to ‘C’. If the two strands were to unwind and detach from each other, Watson and Crick noted, it should be possible to recreate the original strand merely by pairing up each base in the separated strand with its appropriate nucleotide. This idea was called the semi-conservative model of replication.
Other eminent scientists attacked this idea. Max Delbrück was a renowned physicist at the California Institute of Technology who, together with Salvador Luria, had discovered that bacteria resist phage attacks via random mutations. He penned an article arguing that the semi-conservative model could not be correct because too much energy would be required to unwind the two DNA strands.
Delbrück favored a different model, called dispersive replication, in which small chunks of a DNA molecule are broken up, and then matching DNA sequences are synthesized directly in the broken regions to create an intact, double-stranded helix. A third group of scientists favored a conservative replication model, which theorized that the entire DNA molecule is somehow copied without unwinding whatsoever.
Thanks to a particularly innovative experiment devised by two young scientists at Caltech, named Matthew Meselson and Franklin Stahl, Watson and Crick’s semi-conservative model was ultimately vindicated.
It would be relatively simple to figure out how DNA replicates if one could directly observe these molecules. But that was not possible in 1958. Instead, Meselson and Stahl devised a clever experiment, based on spinning molecules quickly in a centrifuge, to test the three models.
Meselson and Stahl’s key insight was to tag DNA strands undergoing replication with heavy atoms, such as nitrogen (N15) that carries an extra neutron. The scientists grew bacterial cells in a growth medium containing this heavy nitrogen, waited for the N15 to incorporate into all of the cells’ molecules, and then quickly transferred the ‘heavy’ microbes into growth media with normal nitrogen.
As the DNA molecules replicated, Meselson and Stahl killed the cells and used a centrifuge to spin down the molecules. As the tubes spin, heavier DNA moves toward the bottom and lighter DNA stays closer to the top. Before the cells replicated their DNA, all of the DNA molecules contained heavy nitrogen. After one round of DNA replication, the DNA strands contained half-heavy and half-light nitrogen atoms (Meselson and Stahl saw two ‘bands’ begin to appear in their centrifuged tubes.) And after two rounds of DNA replication, only one-in-four DNA molecules contained heavy nitrogen, suggesting that the semi-conservative model was correct.
This experiment is renowned for its simplicity and clever approach – it is now called “the most beautiful experiment.” Delbrück was wrong; DNA replication occurs when the two interlocking strands unwind, and each strand is then used as a ‘template’ to remake a double helix.
The Central Dogma (1958)
After publishing his 1953 Nature paper about DNA’s structure, Francis Crick toured the world to lecture on an idea that "permanently altered the logic of biology," according to Horace Judson, author of The Eighth Day of Creation.
During his lectures, Crick would often draw a diagram on the auditorium’s blackboard. His diagram depicted how information flows through living cells; DNA is somehow converted into an intermediate molecule, which Crick called ‘template RNA’, that somehow encoded the amino acids in a protein molecule. Crick correctly predicted the basic details of protein synthesis years before direct experimental evidence had confirmed the existence of mRNA or tRNA.
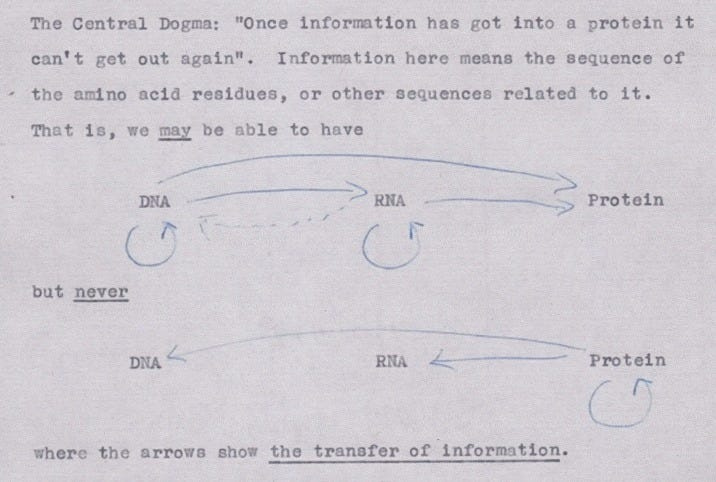
In 1958, Crick adapted his lecture into a published article, called On Protein Synthesis. His target audience was “a general reader rather than the specialist." The article gave two hypotheses to explain the relationship between DNA and proteins, called the Sequence Hypothesis and the Central Dogma.
“The direct evidence for both of them is negligible,” Crick wrote, “but I have found them to be of great help in getting to grips with these very complex problems.”
The sequence hypothesis, in its simplest form, “assumes that the specificity of a piece of nucleic acid is expressed solely by the sequence of its bases, and that this sequence is a (simple) code for the amino acid sequence of a particular protein.” In other words, the bases in a strand of DNA or RNA corresponds to the amino acids in a protein.
The Central Dogma, Crick wrote, “states that once 'information' has passed into protein it cannot get out again.” Stated another way, “the transfer of information from nucleic acid to nucleic acid, or from nucleic acid to protein may be possible, but transfer from protein to protein, or from protein to nucleic acid is impossible.”
This passage marked the first time that the Central Dogma, the defining idea of molecular biology, had been published. But this is not why Crick’s article was so prescient.
In the article, Crick used scattered experimental evidence and anecdotal observations, including the fact that "spermatozoa contain no RNA," to correctly predict that there must be a messenger RNA molecule in the cytoplasm that is produced by "the DNA of the nucleus."
Crick’s astounding ability to theorize was most prominently displayed, though, when he correctly inferred the existence of tRNAs, predicted what they were made of, and explained how they likely became 'charged' with amino acids for protein synthesis.
Molecular biologists knew that proteins were made from 20 amino acids, but most other details of protein synthesis were a mystery. Today, we know that tRNA molecules get 'loaded' with the correct amino acid via the action of specific enzymes, and that this is how a message encoded in a strand of RNA is used by the ribosome to build a protein. But Crick had little evidence for any of this. And yet, in his 1958 paper, he wrote:
"Granted that...[mRNA]...is the template, how does it direct the amino acids into the correct order? One's first naive idea is that the RNA will take up a configuration capable of forming twenty different 'cavities', one for the side-chain of each of the twenty amino acids. If this were so, one might expect to be able to play the problem backwards -- that is, to find the configuration of RNA by trying to form such cavities. All attempts to do this have failed, and on physical chemical grounds the idea does not seem in the least plausible...
Apart from the phosphate-sugar backbone, which we have assumed to be regular and perhaps linked to the structural protein of the particles, RNA presents mainly a sequence of sites where hydrogen bonding could occur. One would expect, therefore, that whatever went on to the template in a specific way did so by forming hydrogen bonds. It is therefore a natural hypothesis that the amino acid is carried to the template by an 'adaptor' molecule, and that the adaptor is the part which actually fits on to the RNA. In its simplest form one would require twenty adaptors, one for each amino acid.
What sort of molecules such adaptors might be is anybody's guess. They might, for example, be proteins...though personally I think that proteins, being rather large molecules, would take up too much space. They might be quite unsuspected molecules, such as amino sugars. But there is one possibility which seems inherently more likely than any other-that they contain nucleotides. This would enable them to join on to the RNA template by the same 'pairing' of bases as is found in DNA, or in polynucleotides.
If the adaptors were small molecules one would imagine that a separate enzyme would be required to join each adaptor to its own amino acid and that the specificity required to distinguish between, say, leucine, isoleucine and valine would be provided by these enzyme molecules instead of by cavities in the RNA. Enzymes, being made of protein, can probably make such distinctions more easily than can nucleic acid."
This paper is a tour-de-force of logical reasoning. It became the focal point, a rallying cry, for molecular biologists seeking to crack the genetic code and resolve the cell’s mysteries. Crick, fortunately, would not have to wait long for his ideas to be vindicated. A ‘template RNA,’ or messenger RNA as it’s now called, was discovered just three years later.
Isolation of messenger RNA (1961)
Messenger RNA was first isolated by two separate research groups in 1961. Their results appeared back-to-back in the 13 May issue of Nature.
At the Institut Pasteur in Paris, the French scientists François Jacob and Jacques Monod had discovered that the enzymes required to break down a sugar in bacterial cells were only made after cells were exposed to that sugar. In other words, cells somehow “process” an external cue and make proteins in response. This marked the discovery of genetic regulation, but also raised a slew of questions.
Among them: How does a cell know which genes to turn on at any given time? Why doesn’t the whole genome “turn on” at the same time? Several answers were proposed. Maybe there is a custom ribosome corresponding to each gene, some said. Or maybe, as Crick had proposed in 1958, there is an intermediate molecule – a “template RNA” – that transmits messages between DNA and proteins.
In 1960, two groups set out to isolate this mystery molecule. The first group rallied around Matthew Meselson’s laboratory at the California Institute of Technology, and included Sydney Brenner and François Jacob. A second group rallied around Wally Gilbert’s group at Harvard, and included James Watson and François Gros, a French biologist who had worked with Jacob.
Both groups turned to a compelling, experimental model: bacteriophages. When E. coli bacteria are infected with a phage, many scientists had noted that the cells stop making their own proteins, and quickly switch over to making the phage proteins. “This system thus provides an ideal model for observing the synthesis of new proteins following the introduction of specific DNA,” Gilbert’s team noted in their 1961 paper.
To isolate messenger RNA, the Caltech scientists grew bacteria in a growth medium with heavy isotopes, much like Meselson had done with Stahl several years earlier to validate the semi-conservative model of replication. These ‘heavy’ bacteria were then infected with phage and immediately transferred into a growth medium with light isotopes. Infected cells were finally lysed open at regular time points and spun down in Meselson’s ultracentrifuges.
The bands that emerged from Meselson’s centrifuges confirmed a few things. First, bacterial cells did not make new ribosomes after they were infected. This observation was evidence against the fact there is a unique ribosome for each gene. Second, the results confirmed that a new type of RNA molecule was swiftly made after phage infection, and that this new RNA quickly attached to existing ribosomes in the cell. This suggested that the DNA in phages was being quickly transcribed into messenger RNA. And third, the bacterial cells began to make phage proteins using their existing ribosomes.
They had discovered messenger RNA. There is an excellent, and much richer, account of this history by the scientific historian, Matthew Cobb.
Mapping a Codon (1961)
Crick’s 1958 paper made a series of predictions about messenger RNA, transfer RNAs, and how a code embedded in a DNA molecule could possibly encode a protein. But one longstanding question in molecular biology had to do with the nature of the genetic code itself. Namely, how do the nucleotides in a strand of RNA encode the amino acids in a protein? What does UAG mean, or GAA, or UUU, or any other codon, for that matter?
The first triplet codon to be mapped to an amino acid was ‘UUU’ to phenylalanine. This connection was made by two young researchers at the National Institutes of Health (NIH) in Bethesda, Maryland.
Heinrich Matthaei was a post-doctoral fellow working in the laboratory of Marshall Nirenberg, a new researcher at the Institutes. The two scientists were interested in the Central Dogma – they had read Crick’s paper – and aimed to understand the connection between RNA and proteins, often by running experiments on cell-free extracts, a liquid made by grinding up living cells in a mortar and pestle. This enabled the two scientists to study cell biochemistry without having to deal with living organisms.
At 3 o’clock in the morning of May 27th, the two scientists took some of these ‘cell guts’ and added a few drops of synthetic RNA with the sequence:
UUUUUUUUUUUUUUUUU
Their concoction was next added to 20 different tubes, each of which held a different amino acid; valine, alanine, glutamine, and so on. One of the tubes contained phenylalanine amino acids that had been labeled with a radioactive isotope.
“The results were spectacular and simple at the same time,” according to a brief history from the NIH. “After an hour, the control tubes showed a background level of 70 counts, whereas the hot tube” – with the radioactive phenylalanine – “showed 38,000 counts per milligram of protein.”
In other words, when the synthetic RNA molecule was added to a tube of phenylalanine amino acids, the cell-free extract began to churn out radioactive peptides. This singular experiment suggested that the nucleotides UUU somehow encode phenylalanine during protein synthesis.
Over the next several years, Nirenberg and other researchers would go on to map all 64 codons, including the codon that signals the start of translation, AUG. Nirenberg shared the 1968 Nobel Prize in Physiology or Medicine.
Cracking the Genetic Code (1961)
The year 1961 was molecular biology’s annus mirabilis. Messenger RNA was isolated for the first time and Nirenberg and Matthaei decoded the ‘meaning’ of the first codon – UUU. Even after those papers were published, though, mysteries remained. Among them: Is the genetic code overlapping or non-overlapping? And is it actually made from doublet, triplet, or quadruplet codons?
A messenger RNA sequence that reads ‘AUGACC’ could be read by the ribosome as 'AUG' and then 'ACC,' or it could be read by the ribosome as ‘AUG’, ‘UGA,’ ‘GAC’, ‘ACC’. The former is a non-overlapping code, and the latter is an overlapping code. Similarly, the code could be read as ‘AU’ and then ‘GA’ and then ‘CC’ if codons were doublets, or ‘AUGA’ and then ‘UGAC’ if they were quadruplets, and so on. Nirenberg and Matthaei’s experiment did not help to answer either of these questions, because their synthetic RNA had a repetitive sequence: UUUUUUUU.
In the waning weeks of 1961, Sydney Brenner, Lesie Barnett, Francis Crick, and R.J. Watts-Tobin used fragmentary experimental evidence and thought experiments to conclude that each amino acid in a protein is encoded by a triplet code, and that the letters in this code do not overlap. Their ideas were published in a paper entitled, “General nature of the genetic code for proteins.”
Their experiments hinged on two things: A bacteriophage, called T4, that infects bacteria, and a particular type of dye, an acridine called proflavin, that precisely mutates DNA by adding or removing a single nucleotide.
Crick, the ever-careful thinker, had a beautiful idea. He decided to take some T4 bacteriophage and then expose it to proflavin, such that the phage lost its ability to make a particular protein. If Crick added one base and then removed one base, using the acridine, he noted that the phage were able to make the protein. But if he used acridine to add two bases, the phage did not make the protein. When three bases were added, the phage made the protein again. From these observations, the scientists argued that the genetic code must use triplets to encode each amino acid. Their takeaway was based on this fragmented, experimental evidence.
Even though the "combination of mutations strongly suggested that the code was based on units of three bases, the experiments could not prove that to be the case – a code using groups of six bases was consistent with the results," wrote Matthew Cobb in a 2021 history of this paper.
Today, we know that there are 64 codons in total, and that codons appear as ‘triplets’ to encode amino acids in a final protein chain. Codons made of six bases “would raise all sorts of problems,” as Cobb notes, “by massively increasing the number of either meaningless or degenerate sequences (there would be 4096 possible combinations of bases, rather than a mere 64).”
As Crick later said: This was “hardly likely to be taken seriously.”
Translation via a Single Ribosome (2008)
aBy 1961, the basic contours of the Central Dogma had been resolved. But that doesn’t mean all work has since abated, nor that the years from 1953 to 1961 are all-encompassing. Linus Pauling at Caltech predicted the main structural motifs of proteins as early as 1951. A ‘stop’ codon that halts protein synthesis was identified in 1965. The ribosome’s structure was solved in 2000, after decades of work, and culminated in the 2009 Nobel Prize in Chemistry.
Today, synthetic biologists continue to expand the Central Dogma using technologies that Francis Crick, in 1958, could only have dreamed of. And yet, the molecular choreography that underlies the Central Dogma continues to surprise. There are far more enzymes and components involved than early molecular biologists ever could have realized. Transfer RNAs carry amino acids to the ribosome, proteins interact with the ribosome to push it off the RNA strand, and dozens of proteins are involved in transcription initiation, elongation, and termination in human cells.
Molecular biologists continue to resolve this complexity today. In a 2008 study, called “Following translation by single ribosomes one codon at a time,” chemists at the University of California, Berkeley studied individual ribosomes as they moved along a single messenger RNA molecule. Their experiment revealed the stochastic starts and stops of a ribosome during translation.
For this experiment, each end of an mRNA molecule was attached to a polystyrene bead. One of the beads was then placed in a laser trap, holding it in place. The middle of the mRNA molecule contained a long loop, which slowly unwound as the ribosome traversed along its length. As the mRNA molecule stretched out, this elongation could be directly measured by measuring the distance between the two beads.
The chemists repeated this experiment several times, and measured the rate at which the mRNA molecule stretched out each time. Their key result was this: Ribosomes do not glide along the mRNA at a steady pace (which would stretch out the molecule in a linear fashion), but rather jump from one codon to the next in time steps of around 0.1 seconds. The ribosome occasionally pauses between jumps. Each ribosome, then, translates a strand of mRNA in a slightly different amount of time.
This experiment is one of thousands that have been applied to study the Central Dogma in the last two decades. Crick’s 1958 article continues to inspire generations of molecular biologists, who have found his ideas to be rich fodder for a lifetime of scientific work. We now know a shocking amount about transcription, translation, and the genetic code; bacteria add about eight amino acids to a protein each second, human cells add about five amino acids in the same length of time, and DNA is transcribed to RNA at a rate of about 40 nucleotides per second.
Conclusion
The ways in which cells process information is a biophysical marvel that has slowly unraveled over the last 70 years. The Central Dogma, and the 7 seminal experiments described in this essay, are the basis for most everything we do in genetic engineering. But there are still many instances in which we, as genetic designers, place a gene into a cell expecting one thing to happen, but observe something entirely unexpected instead. In other words, biology does not always behave as we expect.
Though useful, the Central Dogma is an incomplete way to think of a living cell. DNA is not always transcribed to RNA, and RNA is not always translated into protein. Sometimes RNA goes back to DNA. The only rule in biology is that there are exceptions to every rule. In future posts, we’ll continue our exploration of the Central Dogma and explain many of these exceptions.
Come join us or subscribe for updates.
Contributors: Ben Gordon and Alec Nielsen. Words by Niko McCarty.
Lead image: Marshall Nirenberg poses next to his personalized license plate in 1987. Credit: National Institutes of Health










NICE HISTORY OF DNA..INTERESTING ...
First, I really like this historical analysis. There are always interesting details that get uncovered. I just have one question. If Crick regretted naming it the Central Dogma, why do synthetic biologists feel the need to build on that, when Darwin was also speculating on primordial origins? According to my late professor, Carl Woese, Molecular Biologists believed that the genetic code was a frozen accident, when it was in fact optimized from statistical mechanics (Goldenfeld, 12/2023: https://www.youtube.com/watch?v=l11VDjUXEJI)
https://journals.asm.org/doi/full/10.1128/mmbr.68.2.173-186.2004
https://github.com/hatonthecat/Biology