
David Goodsell is a biochemist who moonlights as a watercolor painter. He’s renowned for his 1,000,000x magnifications of cell interiors — proteins, pathogens, and more. He measures each molecule’s dimensions and paints them true to scale. His style has become almost nostalgic; many science illustrators borrow his visual style whether they realize it or not.
In recent weeks, I’ve been playing around a bit with PyMOL (and other tools) to visualize proteins. I find myself returning to Goodsell’s visuals again and again, perhaps because my brain has been pre-trained to interpret molecular biology through his eyes. Below, I’m sharing a set of PyMOL commands that you can run to view proteins in Goodsell’s style. Then, I explain how to do the same thing using ChatGPT, and close with links to other tools and resources to visualize molecules.
Quick PyMOL Tutorial
Click here to download PyMOL. It’s free for personal use. Then, find the protein you’d like to visualize on the Protein Data Bank. Once you’re ready to go, use these basic commands to render a protein in David Goodsell’s style.
Setup
> set assembly, 1 # Some PDB structures are dimers or larger complexes; this command instructs PyMOL to display only the first “assembly” in the PDB file.
> fetch 4CMP # This four-digit code represents a Cas9 structure from the PDB.
Background and Sticks
> bg_color white # Set the background color.
> as sticks # Display the structure as sticks, rather than balls or lines.
> set stick_radius = 1.7 # Expand stick radii so they appear more cartoonish.
Coloring the Protein
> color lightblue, not org # Setting baseline colors
> color magenta, org
> select RuvC_domain, (chain A and resi 1-60) or (chain A and resi 718-775) or (chain A and resi 909-1099) # The RuvC domain in Cas9 stretches from amino acids 1-60, 718-775, and 909-1099. This command selects those segments and names it as “RuvC_domain”.
> color lightorange, RuvC_domain # Color the selection as orange.
Render the Image
> unset specular # Disables specular reflections in the rendered model.
> set ray_trace_gain, 0
> set ray_trace_mode, 3 # Gives the cartoon-ish style.
> set ray_trace_color, black # Accentuate the shadows.
> unset depth_cue
> ray # Make a high-resolution image.
This may seem like a lot to memorize, but don’t worry. There’s an easier way: ChatGPT.
Modern LLMs are remarkably good at writing PyMOL commands. But you have to give them detailed prompts, or else the output will not be what you want. They can’t intuit your desired visual style unless you spell it out.
I recently prompt OpenAI’s o1 model with:
Write a series of line commands for PyMOL to visualize a protein in the style of David Goodsell. Fetch and style the protein with a PDB ID of 4CMP.
The model returned a couple dozen line commands for PyMOL. Running them step-by-step yields a decent visual approximation.
Some takeaways: ChatGPT doesn’t know which domains to highlight. In this case, it randomly guessed that the RuvC domain corresponds to amino acids 1–100 and colored them light blue. Also, the resulting style wasn’t exactly what I envisioned; I was imagining something more globular and Goodsell-like. But without explicit instructions, the model can’t read my mind. Still, I’m quite pleased with how much o1 can achieve with just two short lines of prompting!
More Resources
Excellent PyMOL video tutorials here.
If you’re looking to make “close-up” images of protein active sites or interactions, for example, I strongly prefer Ross Wilson’s visual style. Full tutorial here.
PyMOL is not the only tool to do this. David Goodsell has also released a Fortran program, called Illustrate, to make non-photorealistic molecule illustrations. It’s free.
CellScape is a Python library for cartoonish protein structure visualizations. GitHub repository here.




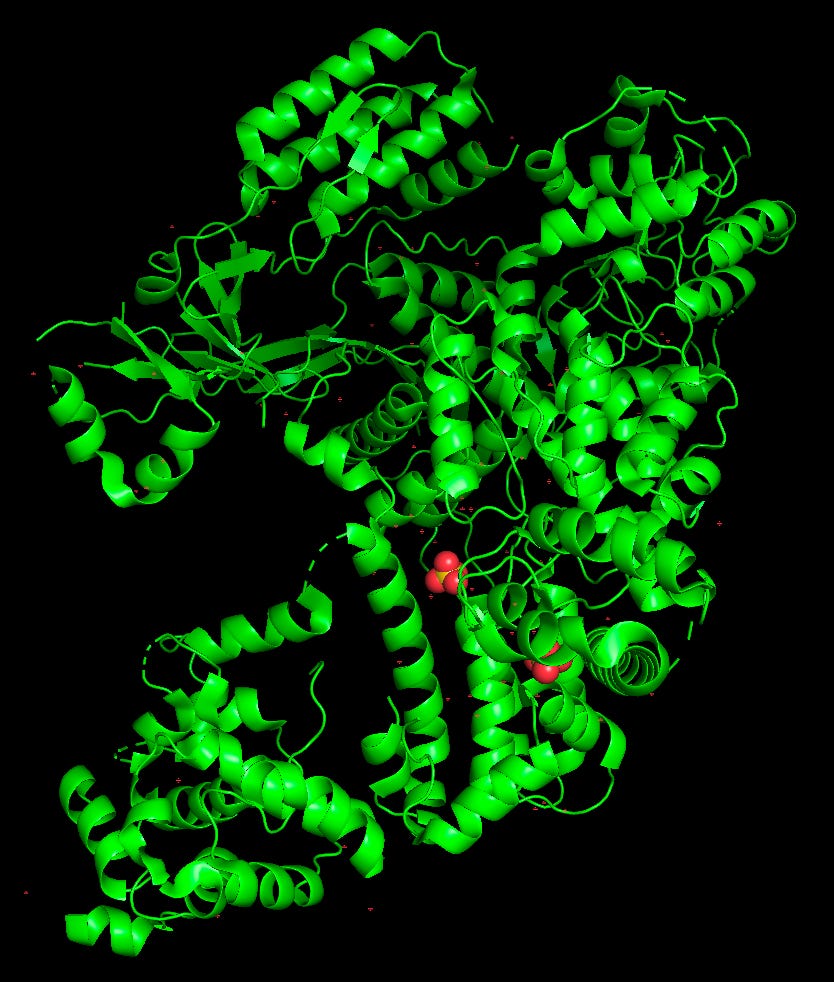

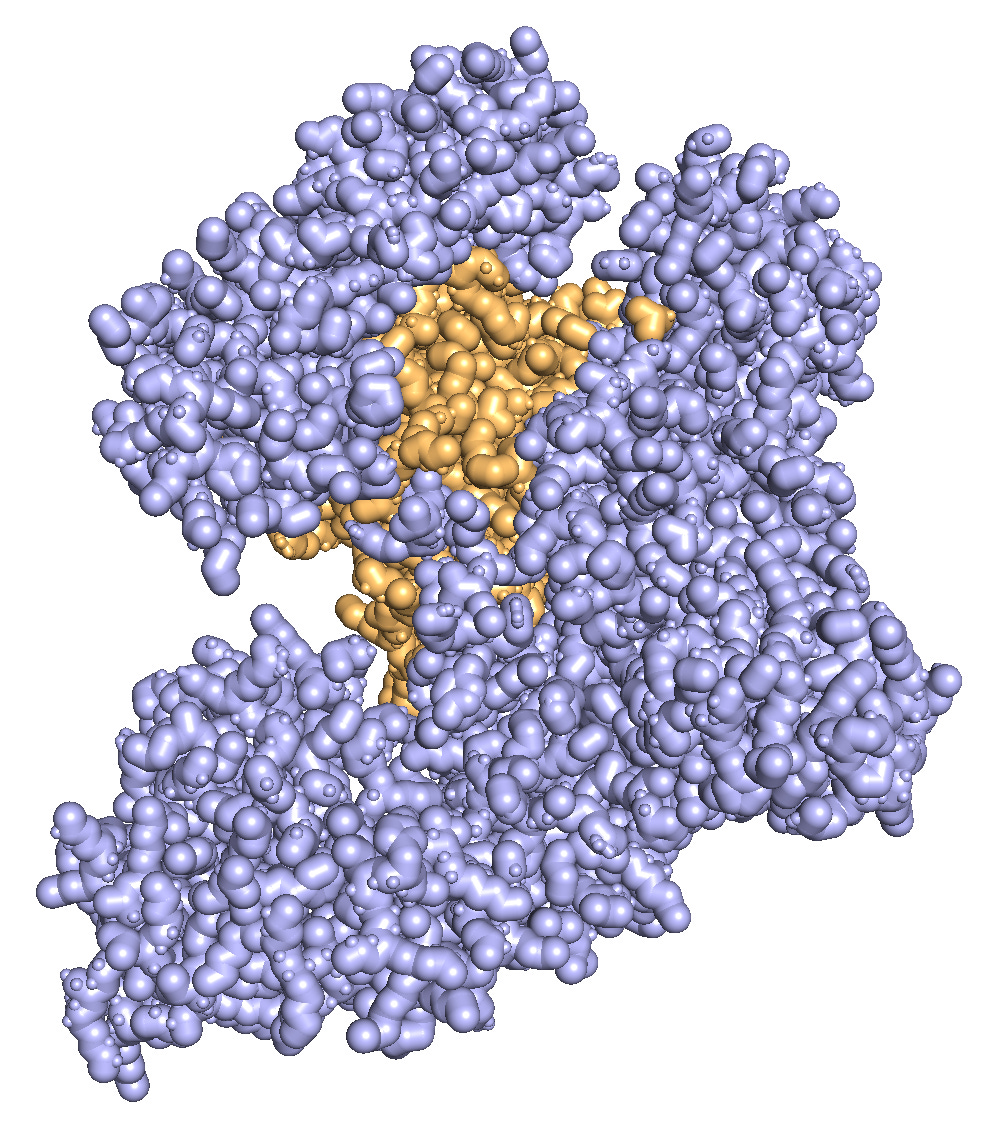
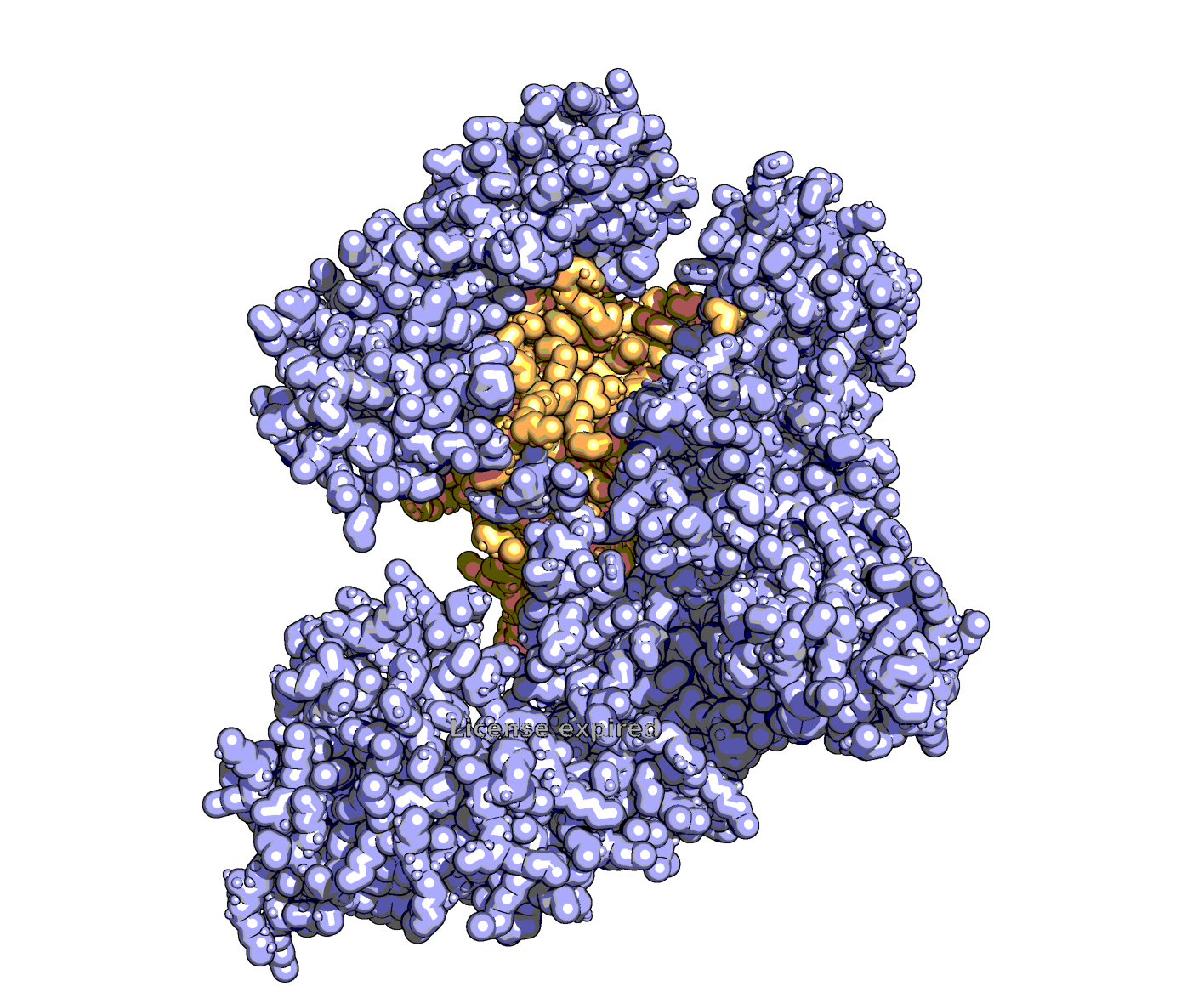
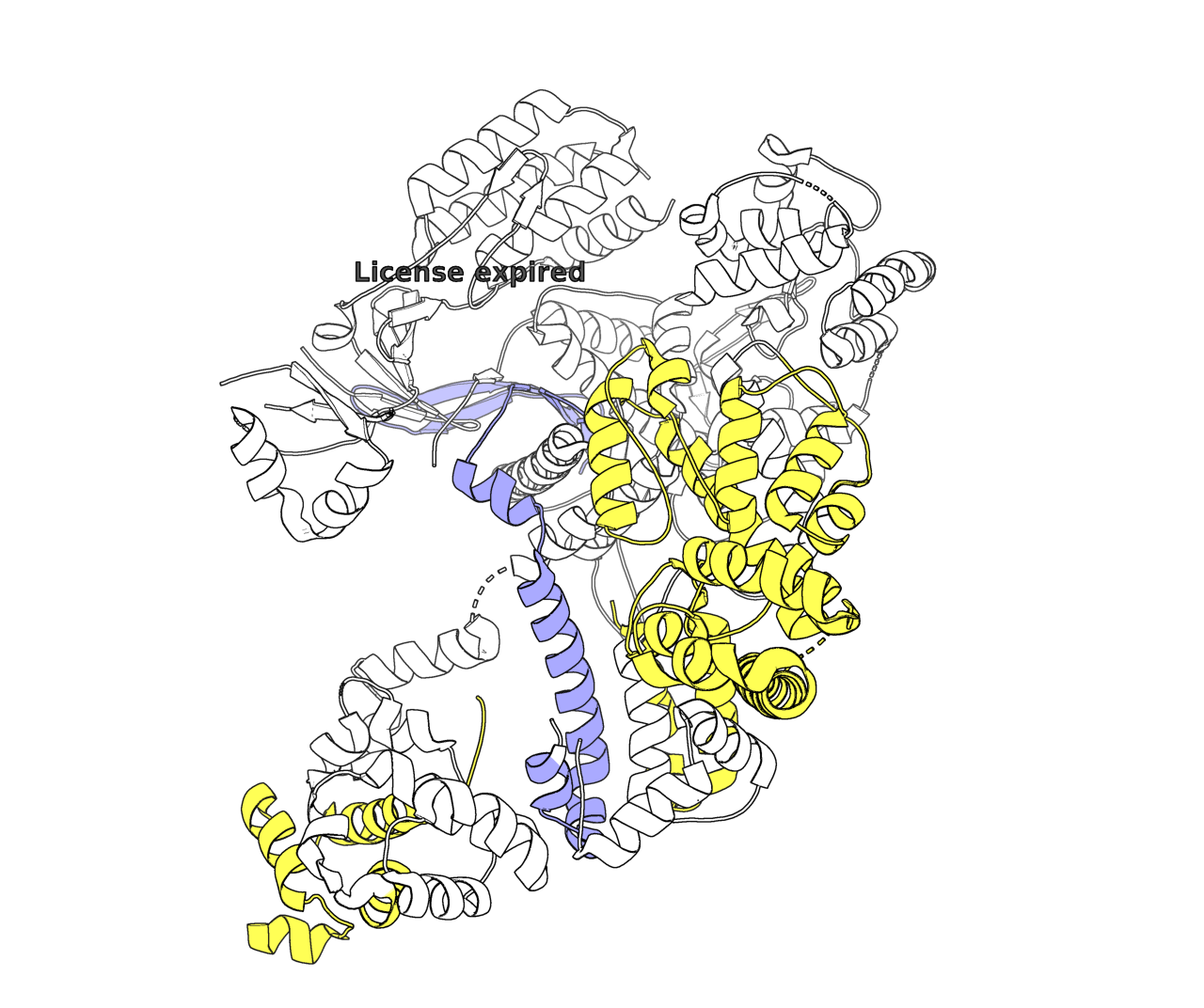
>Click here to download PyMOL. It’s free for personal use. Then, find the protein you’d like to visualize on the Protein Data Bank. Once you’re ready to go, use these basic commands to render a protein in David Goodsell’s style.
Note, pymol is also available as open-source software: https://github.com/schrodinger/pymol-open-source or https://anaconda.org/conda-forge/pymol-open-source
So you can install it and not have to worry about not having a license. The "free for personal use" stuff is only for the prebuilt binaries. Schrodinger is taking advantage of the fact that many biologists don't know how to install stuff from the command line.