Genetic Design in 7 Experiments
We’ve repeatedly touted the idea of genetic design, a research field that aims to apply biophysical insight to compose and layer genetically-encoded functions to achieve cellular behaviors.
If you scratch out the jargon, that means we want to work backwards from an objective — synthetic photosynthesis or the eradication of a cancerous cell in the body — to a DNA sequence that encodes all the biochemistry needed to make that happen.
Genetic design overlaps with synthetic biology in many ways, but the two aren’t the same. Genetic design is deeply rooted in biophysics and mathematics. It’s about understanding how different cellular components can work together to accomplish a desired behavior, and then building it.
Progress in biology is arguably improving faster than any other time in human history, but even advanced models can’t capture all the complexity of a single cell. We cannot fully model biology, and thus our designs often fail. That’s because there is much we don’t understand about life; thousands of genes in a bacterium as simple as E. coli still have unknown functions.
Further complicating matters, cells don’t behave like little machines, despite the metaphors we ascribe to them. Biology is stochastic; probabilistic. Genes are not “switched on” at a steady rate, but fire instead in frenzied bursts of activity. Some proteins behave more like squiggly liquids than solids, or entirely change their function when moved from one environment to another (this is known as moonlighting).
And still, even with this foggy and incomplete view, our ability to do genetic design is rapidly improving. This is the story of the field, as told through seven seminal experiments. It is inspired by The Generalist’s essay on the history of AI.
The lambda OR control system (1985)
Stéphane Leduc, a French chemist, was first to coin the phrase “synthetic biology.” In the early 1900s, he described biology as a series of chemical and physical forces that could be modeled with mathematics, and attempted to make life anew by mixing various chemicals. He was unsuccessful, but did make some pretty sculptures.
Over the last century, scientists have devised new tools to unravel the inner workings of living cells (including DNA sequencing and -omics technologies), and the data they’ve collected suggest that Leduc was, perhaps, ahead of his time. Biology can be modeled using mathematics, and cells — even if not built from scratch — can at least be endowed with new functions.
Early work in genetic design focused on bacteriophage lambda, or lambda phage, a tiny virus that infects E. coli bacteria. This phage can exist in two different states: lysogeny, where it incorporates genetic material into a bacterium, or lytic, where it reproduces and then bursts from the cells in a violent wave.
The switch from one state to the other is controlled by just three genes, and the outcome can be watched with microscopes. Together, these traits make lambda phage an ideal test case to compose mathematical equations and test them in the laboratory.

In a 1985 paper, two biophysicists at Johns Hopkins devised a mathematical model to study lambda phage’s “switching behavior.” Madeline Shea (now a professor at the University of Iowa, and a former mentor of mine) and Gary K. Ackers’ predicted whether a phage would enter the lytic or lysogeny state based on the positions of little DNA sequences within one of the gene’s promoters.
They used statistical mechanics (probability theory to understand how large groups of molecules behave) to calculate how often proteins and strands of DNA came together, and then differential equations to predict whether that would be sufficient to switch from one state to the other.
Calculations were done on a Hewlett-Packard 1000, a $23,900 computer with 768 kilobytes of memory. At the time of its release, these computers “were so fast, they were bound by special US government export restrictions,” according to the HP Computer Museum.
And the Shea-Ackers model worked. It predicted phage behaviors over extended time periods, and later became the mathematical precipice upon which synthetic biology was founded.
Circuit simulation of genetic networks (1995)
Open a biochemistry textbook and flip to a section on metabolism. Peer upon the spaghetti monster diagrams that depict biochemical pathways, in which lines connect rectangles to lines and more rectangles. These diagrams are a chaotic effort to turn a living cell’s complexity into clean lines and digestible iconography.
The lambda phage genome has merely 48,500 bases of DNA, and its life cycle is controlled by just three genes. But much of biology is not like this; it is far messier. Most biochemical networks are built from hundreds of interacting pieces. And, just thirty years ago, there were no tools or frameworks to help us understand the complex dynamics of such large networks.

But then, in the mid-1990s, Harley McAdams and Lucy Shapiro devised a “hybrid modeling approach” to depict genetic networks. They combined “conventional biochemical kinetic modeling within the framework of a circuit simulation.” For perhaps the first time, biophysicists began to sketch out the complex interactions between biochemical “stuff” — DNA, promoters, genes, proteins — as circles and lines. Each biochemical object was then attached to another using the logic gate symbols from electrical engineering, where a triangle means “NOT,” a half-circle means “AND,” and so on.
By modeling biology in a way akin to electrical circuits, it became far easier to build a predictive software for living cells. Each biochemical “thing” was stored as an object, and could be manipulated with a suite of functions to piece together a full biochemical network.
“We are optimistic that libraries of generic object-oriented software models of common genetic mechanisms,” wrote McAdams and Shapiro, “can be developed to provide geneticists the type of user-friendly simulation tools that electrical circuit analysts now take for granted.”
This was a prediction decades ahead of its time.

Early gene circuits (2000)
Much of the history of molecular biology was written by physicists. Erwin Schrödinger popularized a physical view of genetics. Francis Crick co-discovered the structure of DNA and was first to theorize the Central Dogma (he got his start in physics, and later designed acoustic mines for the British army during World War II.) Walter Gilbert studied exclusively physics through his PhD, co-founded Biogen, and developed new DNA sequencing technologies.
The same can also be said for synthetic biology. The field was founded mainly by physicists who modeled biological networks on a computer, took them apart, and rebuilt them anew to endow cells with new behaviors.
In 2000, back-to-back papers appeared in Nature describing early examples of synthetic gene circuits. A Princeton group, made up of a physics Ph.D. student and theoretical physicist, endowed bacteria with a gene circuit, called the repressilator, that made cells flash a green color – on and off – at regular intervals. A Boston University group, led by another physicist, introduced the toggle switch on the same day.
The toggle switch is made from just two genes; let’s call them A and B. Each gene encodes a protein that represses the other, and can itself be controlled by an external factor. Protein A is switched “off” by a small molecule, for example, and protein B is switched “off” by heat. This simple co-repressive design makes it possible to control “toggle” cells between two states: Add some heat and protein A dominates. Add the small molecule, and the opposite happens.
The repressilator was made from three genes, each repressing another. Gene A encodes a protein that blocks Gene B, which encodes a protein that blocks Gene C, and so on, in a perpetual inhibitory loop (in electrical engineering, this is called a ring oscillator). Each protein is also attached to a small peptide that speeds up its degradation, so that the cycle can continue to progress. This causes each protein, in the cells, to come up-and-down in its levels at a predictable frequency; 150 minutes.

These papers laid the foundation for synthetic biology. They showed that it’s possible to use simple mathematical models to build DNA sequences that endow living cells with new behaviors. A third paper, published just a few weeks later, reported a third synthetic gene circuit made from just one, self-repressing gene.
And from there, explosive growth ensued.
Programmed pattern formation (2005)
Technologies tend to follow a predictable arc. The first demonstration is usually primitive. But vigilant people take note, tinker around with the new technology, and gradually push it to new extremes. This happened with CRISPR gene-editing: After the early papers, hundreds of scientists quickly pivoted into the field, discovered other gene-editing proteins, made dozens of computational tools, and built an entire ecosystem of genetic engineers.
The early genetic circuit papers may have initiated synthetic biology, but it was other scientists who pushed designs to new extremes — more equations, more biochemical components, more computer simulations.
The first synthetic biology conference was held at MIT in 2004. In the following year, an electrical engineering team at Princeton, led by Ron Weiss, described a genetic circuit that programmed groups of cells to form patterns in a petri dish. This paper was historically notable for a few reasons.
Whereas early genetic circuits were confined to individual cells, this version added a “communication” component; all the cells could talk to each other. The design was also incredibly fine-tuned. It required 100 experiments and 2,000 computer simulations to arrive at a solution, and only 30% of the designs produced the pattern.
First, the researchers placed ‘sender’ cells, programmed to make a small molecule called AHL, in the middle of a petri dish. The molecule diffuses out to nearby cells, called ‘receivers,’ that detect the AHL. Specifically, the molecule latches onto a protein called LuxR. When this happens, LuxR “activates” two branches of a circuit, each of which regulates a green fluorescent protein output.
Now here’s the clever bit. When receiver cells sense either very little or a lot of AHL (which happens at short and long distances from the senders), then the signals from the two circuit branches "clash": one branch is on and one is off. This shuts off expression of the fluorescent output.
But when the receiver cells are located at an intermediate distance, the signals traveling through the two branches of the circuit match, triggering the cells to glow green. This interplay of proteins, and a clever circuit architecture, creates a band detector that causes cells to form a glowing ring pattern in a petri dish.
This paper is one of the earliest demonstrations of true genetic design. Mathematical equations gave way to a computational model, which was then used to simulate thousands of designs until hitting upon those that could coax cells to perform an intricate, finely-balanced behavior.
Synthetic Biology Open Language (2009)
In 1886, an American mathematician, named Charles Sanders Peirce, described how logical operations — AND, NOR, OR, and so on — could be performed by “electrical switching circuits.” Prior to this letter, addressed to one of his former students, engineers had built logic machines with purely mechanical mechanisms.
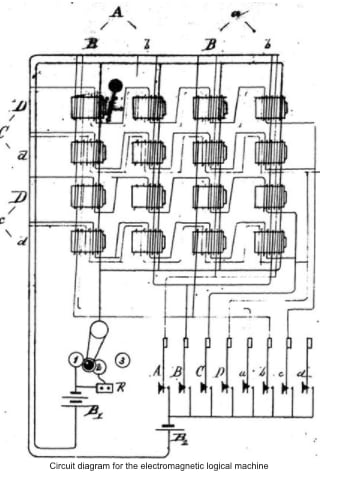
For the next hundred years, logicians and engineers described electrical circuits using all kinds of notations and symbols. Many of these symbols — such as the two parallel lines that represent a capacitor — were adopted because the symbol was representative of the device itself; a capacitor accumulates charge on two surfaces placed near one another. But there were no standards; only norms.
In 1984, though, the Institute of Electrical and Electronics Engineers implemented a standard set of “graphic symbols for logic functions.” The manual includes dozens of little symbols and rules to ensure that an electrical circuit described in a Chinese paper could be immediately recreated by an American academic. Standard notation transcends language barriers. They make it possible to do industrial design and rapidly recreate, reconfigure, and prototype new designs.
But all of this was missing in biology, until about 2009, when a group of 76 people across 37 organizations came together to develop the Synthetic Biology Open Language, or SBOL. The group introduced dozens of symbols to depict various DNA sequences, including promoters (depicted as arrows), translation start sites, and even protein degradation tags.

“Such standards could enable synthetic biology companies to offer catalogs of devices and components by means of computer-readable data sheets, just as modern semiconductor companies do for electronics,” the authors wrote. And they further envisioned a future in which these tools would “enable a synthetic biologist to develop portions of a design using one software tool, refine the design using another tool, and finally transmit it electronically to a colleague or commercial fabrication company.”
Design of individual genetic parts (2009)
In 1957, a General Electric employee named Patrick Hanratty developed PRONTO, the first numerical control programming software. It was used to control machining tools; telling them where to cut, how deeply, and so on.
Three years later, a PhD student at MIT, named Ivan Sutherland (who was working with Claude Shannon, the “father of information theory”) created the first true graphical, computer-aided design program. Called Sketchpad, it made it possible to draw shapes on a screen, adjust them, and export them to be reused by others. It laid the entire foundation of graphical-user interfaces.
Today, CAD tools are vastly more complicated and powerful. Many electrical engineers design circuit boards using AutoCAD Electrical, which has built-in tools to pick, say, wires that have continuous load correction and can operate at high temperatures. In other words, modern CAD tools will help you pick the right parts for a job.
This is missing in biology. About 15 years ago, there were no software tools to design biological sequences with predictable outputs, which could then be compiled into more complex genetic circuits. The Ribosome Binding Site (RBS) Calculator was really the tip of the iceberg.
In cells, ribosomes are the big proteins that make other proteins. In bacteria, they latch onto a small snippet of messenger RNA, called the RBS, and begin translation. This short sequence plays a large role in how often a piece of RNA is translated into proteins. A ‘weak’ RBS coaxes ribosomes to make a little bit of protein, whereas a ‘strong’ RBS coaxes them to make a lot. The RBS Calculator took the biophysical and thermodynamic equations describing the interactions between ribosomes and RNA molecules, and packed them into a web tool that could design new RBS sequences.

If a specific amount of protein is needed, the tool will suggest an RBS design that is accurate within a variance factor of 2.3. In other words, if you were to target a protein production rate of 100, the RBS Calculator outputs sequences that achieve a range between 43 and 230.
This tool matters because it directly outputs a DNA sequence based on a need. And, in engineering, actionability matters. It’s one thing to publish a mathematical equation, and quite another to use it to build a software tool that others can use. Simplicity and actionable results are what will make genetic design blossom into a field where anyone, anywhere, can design cells with custom behaviors on a computer.
Automated design of complex gene circuits (2016)
Not that long ago, genetic design was made up of miscellaneous and one-off tools, like the RBS calculator, that were useful but separated. A biologist could design an RBS sequence, or sketch out circuit designs using standard notations, but there still wasn’t really anything that could put all these features together, into one software package.
“Genetic circuit design automation,” a 2016 Science paper, marked a seminal shift. Developed by teams at MIT and Boston University, it described a programming language to design entire logic circuits made from DNA. In the spirit of Ivan Sutherland or AutoCAD, it suddenly made it possible to go from ad hoc designs to all the DNA instructions needed to create a genetic circuit that “functioned as specified.”
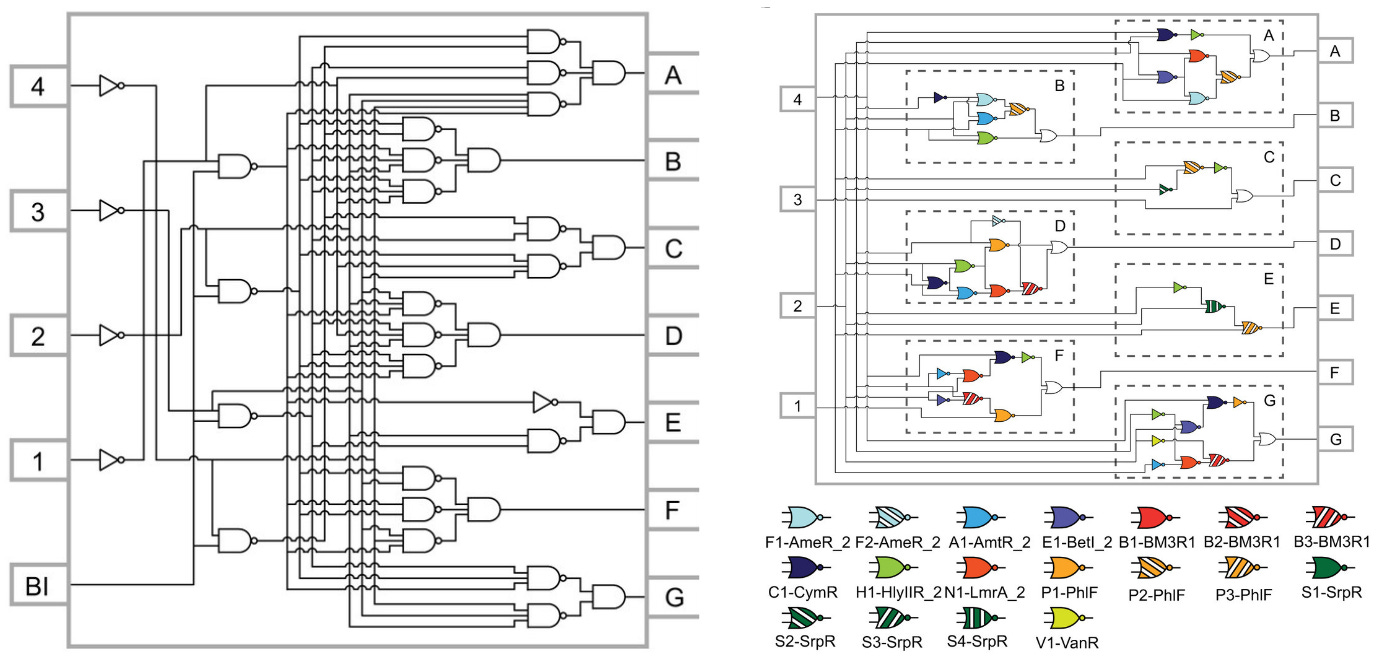
The programming language, called Cello, inspired the technological foundation of Asimov. At the time of its publication, Cello was used to design the most complex genetic circuits ever built — ten proteins that work together to encode complex logic functions — with a 75% first-try success rate. In other words, Cello directly outputs a DNA sequence, based on a user specification, that works as expected three-quarters of the time.
In a series of follow-up papers, Cello was used to build sequential logic, a form of circuit that in real computers underpins memory, timing, and feedback. And, in a separate paper, it was used to program E. coli bacteria “to function as a digital display,” by encoding an entire electronic chip, across multiple strains, in 76,000 bases of DNA.
Genetic design is getting better, fast. A couple decades ago, the earliest synthetic gene circuits were made from two or three genes. Now, genetic circuits are designed on computers and contain dozens of individual parts. If history is any indication, then we may be able to design new organisms, with entirely distinct biochemistries, in forty years. But we’ll first have to study biology as an intricate whole, and understand how our own designs fit into the context of lifeforms that have evolved, without us, over billions of years. Time will tell.
...
Come join us or subscribe below for updates.


.png)